The use of PCR to quantify MRD has been inhibited in North America owing to lack of insurance due to concern by the FDA that the amplification efficiency (AE) of patient-specific primers is unknown and the method is unstandardized. AE is conventionally derived from the slope of the regression line between Ct and the log of input, but the results of this technique are highly variable. Quantification of MRD by PCR is widely used in Europe and slopes corresponding to AE between 80% and 110% have been regarded as acceptable by the EuroMRD consortium.
We have used the Ct resulting from amplification of one copy of a target molecule to provide a measure of AE. The mean Ct from one copy (1.Ct) is determined by depositing rearranged IGH or TCR targets into 24 microplate wells and using limiting dilution and Poisson statistics to identify wells arising from one copy. The method is calibrated by performing multiple accurate estimations of AE, determining them from the slope of the regression line between Ct and logarithm of input DNA. Calibration provides a reference AE, the corresponding reference 1.Ct and the number of amplicons at the PCR threshold (Nt). With this information, the AE of a test primer can be readily determined from its 1.Ct since AE = Nt1/1.Ct - 1.
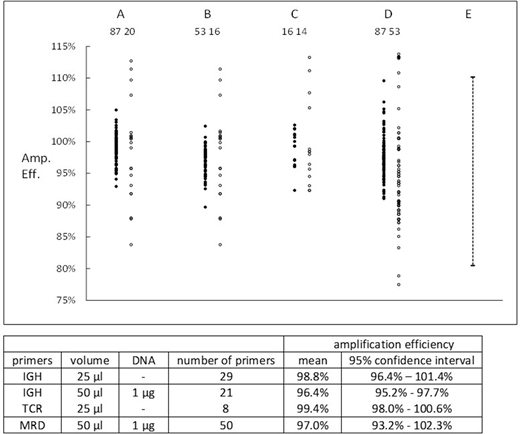
Calibration for IGH primers was performed using the regression lines for each of 20 primers and 10 replicates of a reference primer; calibration for TCR primers used 2 replicates for each of 7 primers. Nt for both IGH and TCR was 1.1 x 1011. Amplification efficiencies of patient-specific primers which had been used for MRD assays were determined either from 1.Ct estimations or regression line slopes and results are shown in the Figure. Closed symbols are 1.Ct results, open symbols are slope results, and the numbers are the number of estimations. Columns: A - IGH primers, 25 µl volume / well; B - IGH primers, 50 µL volume and 1 µg of DNA / well; C - TCR β/γ - 25 µl volume / well; D - retrospective data from MRD assays, 50 µL volume and 1 µg of DNA / well; E - EuroMRD limits for acceptability.
Each set of data showed that use of 1.Ct to determine AE was significantly (p < 0.01) more precise than use of slope. Analysis of variance was performed on the 1.Ct results. Intra-individual variation contributed the majority of variance for all 4 groups. For groups A and D there was significant (p < 0.01) differences between AE of individual primers. The means and 95% confidence limits for individual primers are shown in the Table and indicate that the AE of individual primers was close to 100% and nearly always > 95%.
The MRD primers are all long with a Tm > 69 oC. However, 5 short primers, with a Tm of 60 - 62 oC also amplified efficiently, with a mean AE of 99.1%.
Amplification protocols in other laboratories may differ from those used in our laboratory. To enable standardisation, we have developed a plasmid containing an IGH reference insert. The primers directed to this insert have an AE of 98.8% (95% CI 98.3% - 99.3%) which is constant over an annealing temperature range of 60 - 72 oC.
Although the primers finally used for MRD assays amplified at close to 100%, impaired amplification of MRD candidate primers occasionally occurred due to phenomena such as: an annealing temperature being too high or too low; point mutations in the target sequence; an inhibitory factor in the sample. However , such phenomena were readily identified and eliminated during workup of the primer and sample.
We conclude
1. Patient specific MRD primers amplify at an efficiency close to 100%.
2. Efficiency can be simply and accurately measured.
3. Estimation of MRD by PCR can be a standardised technique.<
Morley:Monoquant:Consultancy, Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees, Patents & Royalties, Research Funding.Latham:Monoquant P/L:Current equity holder in private company, Research Funding.Hughes:Monoquant P/L:Patents & Royalties, Research Funding.Budgen:Monoquant P/L:Patents & Royalties, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.